GFN 19 Found!  |
|
On 13 April 2024, 03:42:53 UTC, PrimeGrid's Generalized Fermat Prime Search found the Mega Prime:
8630170^524288+1
The prime is 3,636,472 digits long and will enter “The Largest Known Primes Database” ranked 11th for Generalized Fermat primes and 83rd overall.
The discovery was made by Antonio Lucendo (Trotador) of Spain using a dual CPU AMD EPYC 7B13 64-Core Processor @ 2.20GHz with 173GB RAM, running Ubuntu 22.04.3 LTS. This computer took about 4 hours, 45 minutes to complete the probable prime (PRP) test using Genefer22. Antonio Lucendo is a member of the XtremeSystems team.
The PRP was confirmed prime on 14 April 2024 by an AMD Ryzen 9 7950X3D @ 4.2GHz, running Debian 12.5. This computer took about 26 hours, 11 minutes to complete the primality test using LLR.
For more details, please see the official announcement.
|
|
20th BOINC Workshop May 29-31  |
The registration is now open for the 2024 BOINC Workshop -- this year in person, at CERN.
https://indico.cern.ch/event/1379525/overview
Please register if you plan to attend.
|
|
Aliquot sequence 28^95 has terminated!!!  |
Aliquot sequence 28^95 has terminated!!!

|
|
Electrical work  |
On Thusday 23/4/2024 there are planned some electrical works in my area from 9:00 to 13:30 with power that probably will be tuned off too.
So the server could be not reachable during that times due to loss of internet connection.
|
|
Processing of the square # 36 completed  |
Dear participants, processing of the square # 36 is fully completed! The square:
0 1 2 3 4 5 6 7 8 9 A B
1 2 0 4 5 3 8 B 9 6 7 A
6 8 9 A 7 B 1 5 2 0 4 3
5 3 4 1 2 0 B 9 A 7 8 6
9 6 8 B A 7 0 4 1 2 3 5
B A 7 8 9 6 3 0 4 5 2 1
A 7 B 9 6 8 4 1 5 3 0 2
4 5 3 0 1 2 7 8 B A 6 9
7 B A 6 8 9 5 2 3 4 1 0
3 4 5 2 0 1 A 6 7 B 9 8
8 9 6 7 B A 2 3 0 1 5 4
2 0 1 5 3 4 9 A 6 8 B 7
has 575323340 orthogonal mates which puts it on the 24th place of the rating which include now 5678 positions.
Now "high part" of spectra of ODLS-12 looks like this (square # 36 marked by red):
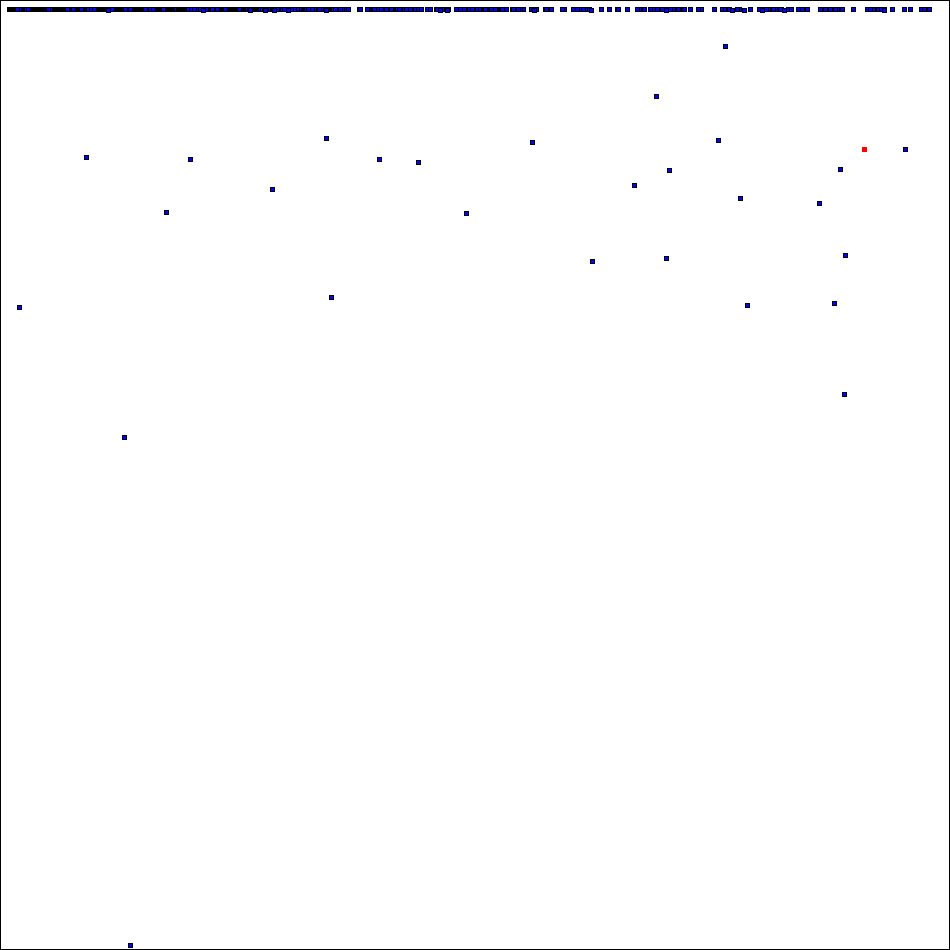
Thank you for project attention, support and donation of CPU time!
|
|
Processing of the square # 35 completed  |
Dear participants, processing of the square # 35 is fully completed! The square:
0 1 2 3 4 5 6 7 8 9 A B
1 2 0 4 5 3 8 9 A B 6 7
5 3 4 0 1 2 9 8 B A 7 6
B 7 9 8 A 6 5 1 3 2 4 0
A 6 8 9 B 7 1 5 2 3 0 4
6 8 A B 7 9 2 3 0 4 1 5
2 0 1 5 3 4 A B 6 7 8 9
4 5 3 2 0 1 7 6 9 8 B A
3 4 5 1 2 0 B A 7 6 9 8
9 B 7 6 8 A 4 0 5 1 3 2
7 9 B A 6 8 3 2 4 0 5 1
8 A 6 7 9 B 0 4 1 5 2 3
has 549717331 orthogonal mates which puts it on the 24th place of the rating which include now 5665 positions.
Now "high part" of spectra of ODLS-12 looks like this (square # 35 marked by red):

Thank you for project attention, support and donation of CPU time!
|
|
Server crash  |
|
The server crashed due a memory leak. The database was corrupt so restored from todays backup, it was expected but need to investigate which was causing this. wanted to fix this in 3d but came earlier.
|
|
base S165 Megaprime  |
[P3D] Crashtest found a member of the team Planet 3DNow! found a megaprime for base S165
The prime 1486*165^497431+1 has 1.103.049 digits and entered the TOP5000 in Chris Caldwell's The Largest Known Primes Database. 1k is left
|
|
Lightning bolt  |
There was a lightning bolt on 1/4/2024 in the night that damage (e.g. they broke) many electrical devices in the area (1Km^2).
Internet connection were restored only in the morning (as provider is at 200m distance of the server and it was damaged too).
Even if the server is behind a power supply, it get in non operational state (e.g the data disk was disconnected), maybe due to the heavy electrical current it passes onto the battery power supply.
Every disk of the server were un-mount from it and tested in another machine and all data were ok, even if the server did not boot properly.
Now it is running in emergency mode (e.g. it need a manual start up, so if it restart from eventual current blackout, it will no boot) and I will monitoring if all is going right.
During the weekend, I will work on fix it, so the server will be disconnected for some times during the day.
Remember that if the internet connection is on and the server is off, you can see some message here: http://boinc-status.multi-pool.info/ about his state.
Please, remember to set up a backup BOINC project in case of emergency. Thanks.
|
|
base R2 2nd conjecture 2 Megaprimes  |
Science United found a megaprime for base R2 2nd conjecture.
The prime 625783*2^7031319-1 has 2.116.644 digits and entered the TOP5000 in Chris Caldwell's The Largest Known Primes Database.
Hoshione, a member of the team SETI.Germany found a megaprime for base R2 2nd conjecture.
The prime 580633*2^7208783-1 has 2.170.066 digits and entered the TOP5000 in Chris Caldwell's The Largest Known Primes Database.
|
|
Part 3 of the 10^21 search has started  |
Part 2 of the 1021 search will be finished, for the most part, today. There are still ~2000 unfinished WUs which can take up to a couple of weeks to complete, depending on individual participants processing them.
Part 3 of the search will look for amicable pairs where the smaller member of the pair has its largest prime factor between 1011 and 1014:
m=m1*p, 1011 < p < 1014
There are too many primes larger than 1014 (all primes up to 5*1019 need to be checked), so they will require a different approach and a new application when the time comes.
Most of the participants have already started getting part 3 WUs. The new work units will require ~2.5-2.7 GB RAM on CPU, and the same amount of RAM on GPU (for GPU applications), so the minimum requirement for GPUs will be 3 GB RAM.
Update April 9th, 2024: I found a bug in OpenCL applications, which resulted in many false negatives (around 12.5%). The bug was in the new code that I added for the part 3, so the previous search is unaffected. Version 3.09 has this bug fixed, but I had to restart the part 3. Luckily, we were only a couple of weeks in.
|
|
BOINC Needs Votes at a UN Upcoming Forum  |
BOINC is a finalist for an notable award, and needs your vote (*by Sunday)
The World Summit on the Information Society (WSIS) (https://www.itu.int/net4/wsis/forum/2024) is a United Nations-sponsored initiative aimed at harnessing the potential of information and communication technologies to build inclusive and equitable information societies worldwide. BOINC has been nominated for a prize at the 2024 forum (https://www.itu.int/net4/wsis/stocktaking/Prizes/2024), and has passed initial hurdles; the next and last step ("Phase 3") requires public votes. The award would be a very nice boost and validation for BOINC and all our projects; if we can get our communities to vote, we should have a decent shot at this point...
Voting is pretty simple, takes just a few minutes; instructions are here: https://docs.google.com/document/d/1x9Xi3tq7Y9dlDD0Xb0Ul0yYCXct5pDqIqssqARxvrXg/edit.
(*The deadline for votes is Thursday April 04 (23:00 UTC+02:00))
|
|
BOINC is a finalist for an notable award, and needs your vote (*by Sunday)  |
The World Summit on the Information Society (WSIS) is a United Nations-sponsored initiative aimed at harnessing the potential of information and communication technologies to build inclusive and equitable information societies worldwide. BOINC has been nominated for a prize at the 2024 forum, and has passed initial hurdles; the next and last step ("Phase 3") requires public votes. The award would be a very nice boost and validation for BOINC and all our projects; if we can get our communities to vote, we should have a decent shot at this point...
Voting is pretty simple, takes just a few minutes; instructions are here.
(*The deadline for votes is Sunday: 31 March 2024, 23:00 UTC+02:00)
|
|
BOINC Needs Votes at a Upcoming UN Forum  |
BOINC is a finalist for an notable award, and needs your vote (*by Sunday):
Context: The World Summit on the Information Society (WSIS) is a United Nations-sponsored initiative aimed at harnessing the potential of information and communication technologies to build inclusive and equitable information societies worldwide. BOINC has been nominated for a prize at the 2024 forum, and has passed initial hurdles; the next and last step ("Phase 3") requires public votes. The award would be a very nice boost and validation for BOINC and all our projects; if we can get our communities to vote, we should have a decent shot at this point...
Voting is pretty simple, takes just a few minutes; instructions are here.
(*The deadline for votes is Sunday: 31 March 2024, 23:00 UTC+02:00)
|
|
Aliquot sequence 21546 has terminated!!!  |
The sequence 21546 has terminated with the amicable pair 1184/1210. It is the smallest discovered terminated sequence since 2012 and the first terminated sequence below 10e5 since 2020. The largest term of this sequence reaches 153 digits which is a new record.

|
|
Planned power maintenance outgage  |
We have a planned network outage in the CAMK building scheduled for Saturday, March 23rd, 2024.
The outgage start on 7pm CET and finish on Monday, 25th, 8am.
|
|
Formula BOINC 2024-03-21 06:00 UTC - 2024-03-24 06:00 UTC  |
|
The challenge is running. Good luck everyone!
|
|
Processing of the square # 34 completed  |
Dear participants, processing of the square # 34 is fully completed! The square:
0 1 2 3 4 5 6 7 8 9 A B
1 2 3 4 5 0 8 6 A B 9 7
5 0 1 2 3 4 7 B 6 A 8 9
7 6 8 A 9 B 1 0 2 4 3 5
6 8 A 9 B 7 2 1 3 5 4 0
B 7 6 8 A 9 0 5 1 3 2 4
8 A 9 B 7 6 3 2 4 0 5 1
9 B 7 6 8 A 5 4 0 2 1 3
A 9 B 7 6 8 4 3 5 1 0 2
2 3 4 5 0 1 A 8 9 7 B 6
4 5 0 1 2 3 B 9 7 8 6 A
3 4 5 0 1 2 9 A B 6 7 8
has 610170218 orthogonal mates which puts it on the 22th place of the rating which include now 5651 positions.
Now "high part" of spectra of ODLS-12 looks like this (square # 34 marked by red):
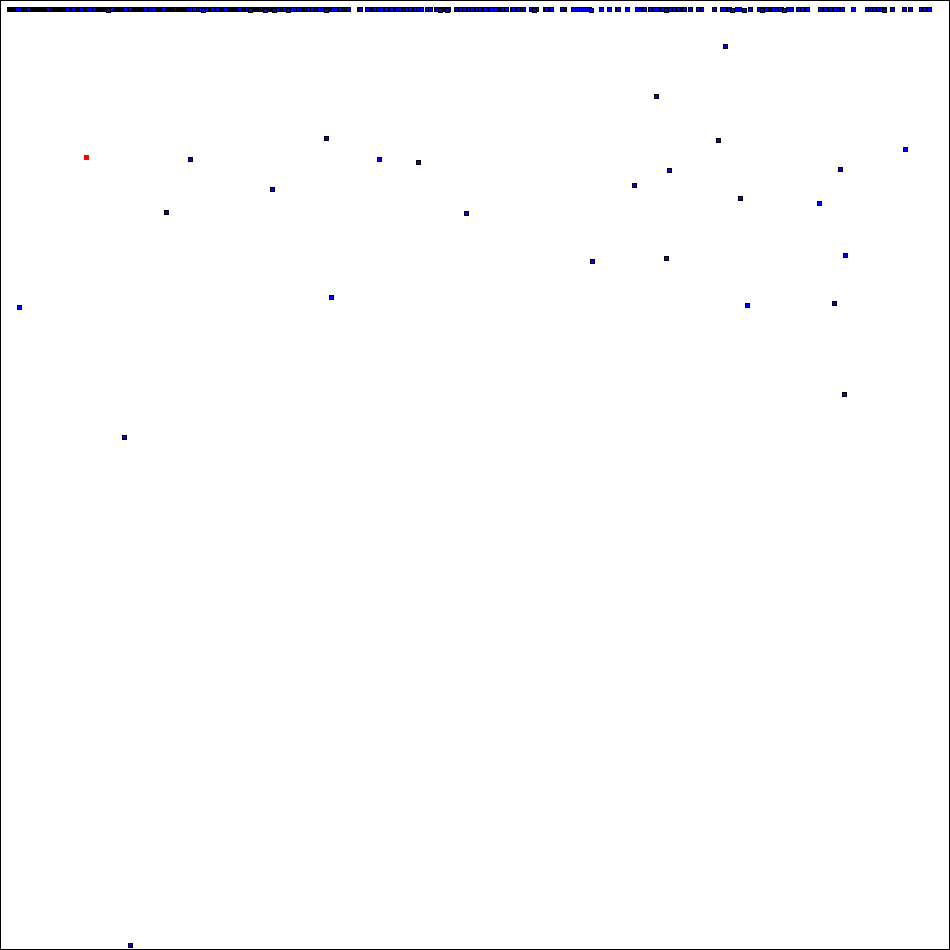
Thank you for project attention, support and donation of CPU time!
|
|
base R815 Magaprime / proven  |
magic_sam, a member of the team Gridcoin found a megaprime for base R815.
The prime 8*815^559138-1 has 1.627.740 digits and entered the TOP5000 in Chris Caldwell's The Largest Known Primes Database. With this find it also has proven the base!
|
|
Aliquot sequences 2772150 and 2771604 have terminated!!!  |
Aliquot sequences 2772150 and 2771604 have terminated!!!


|
|
